mindmap
root((Feature engineering))
Feature transformation
Handling missing values
Imputation
Remove observation
Mean replacement
Median replacement
Most frequest categorical
Handling categorical values
One-hot-encoding
Binning
Handling outliers
Outlier detection
Outlier removal
Feature scaling
Standardization
Normalization
Feature construction
Domain knowledge
Experience
Feature selection
Feature importance
Feature extraction
Feature engineering
This is my notes on feature engineering. The idea is to populate this article with all possible relevant knowledge I can get on my hands on and then use it as a reference in the future. Plan is also to use what I Study and note once, use many times.
import polars as pl
input_data_path = f"../data/iot/iot_telemetry_data.parquet"
df_original = pl.read_parquet(input_data_path)
df_original.head(1)
shape: (1, 9)
| ts | device | co | humidity | light | lpg | motion | smoke | temp |
|---|---|---|---|---|---|---|---|---|
| f64 | str | f64 | f64 | bool | f64 | bool | f64 | f64 |
| 1.5945e9 | "b8:27:eb:bf:9d:51" | 0.004956 | 51.0 | false | 0.007651 | false | 0.020411 | 22.7 |
1. Feature transformation
Importing modules
import duckdb
import polars as pl
import seaborn as sn
import pandas as pd
import matplotlib.pyplot as pltReading data
# ts is in epoch time format so converting it to timestamp
# rounding values for temperature and humidity
# converting temperature from farhaneit to celsius
df_raw = duckdb.sql(
f"SELECT ts, to_timestamp(ts) AS timestamp, device, temp,ROUND((temp - 32) * 5.0 / 9, 4) AS temp_c, ROUND(humidity, 4) AS humidity, lpg, smoke, light FROM '{input_data_path}'"
)Exploring the data
The seven questions to get insight into the data
How big is the data?
# Converting to polars to easy statistics and exploration
df_original = df_raw.pl()
df_original.shape(405184, 9)Imputation / Handling missing values
This dataset is quiet clean, there are no missing data in the input data in any feature. Docs Reference
df_original.null_count()
shape: (1, 9)
| ts | timestamp | device | temp | temp_c | humidity | lpg | smoke | light |
|---|---|---|---|---|---|---|---|---|
| u32 | u32 | u32 | u32 | u32 | u32 | u32 | u32 | u32 |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Handling categorical variables
Outlier detection
Feature scaling
Datetime transformation
The ts column is in unix seconds, which we will convert to timestamp.
df_original = df_original.with_columns(pl.from_epoch(pl.col("ts"), time_unit="s").alias("timestamp"))
df_original.head(2)
shape: (2, 9)
| ts | timestamp | device | temp | temp_c | humidity | lpg | smoke | light |
|---|---|---|---|---|---|---|---|---|
| f64 | datetime[μs] | str | f64 | f64 | f64 | f64 | f64 | bool |
| 1.5945e9 | 2020-07-12 00:01:34 | "b8:27:eb:bf:9d:51" | 22.7 | -5.1667 | 51.0 | 0.007651 | 0.020411 | false |
| 1.5945e9 | 2020-07-12 00:01:34 | "00:0f:00:70:91:0a" | 19.700001 | -6.8333 | 76.0 | 0.005114 | 0.013275 | false |
What is the date range of the data collected?
The time range of the dataset is
print(df_original.select(pl.col(['timestamp', 'ts'])).describe())shape: (9, 3)
┌────────────┬────────────────────────────┬───────────────┐
│ statistic ┆ timestamp ┆ ts │
│ --- ┆ --- ┆ --- │
│ str ┆ str ┆ f64 │
╞════════════╪════════════════════════════╪═══════════════╡
│ count ┆ 405184 ┆ 405184.0 │
│ null_count ┆ 0 ┆ 0.0 │
│ mean ┆ 2020-07-16 00:06:56.798528 ┆ 1.5949e9 │
│ std ┆ null ┆ 199498.399262 │
│ min ┆ 2020-07-12 00:01:34 ┆ 1.5945e9 │
│ 25% ┆ 2020-07-14 00:20:00 ┆ 1.5947e9 │
│ 50% ┆ 2020-07-16 00:06:28 ┆ 1.5949e9 │
│ 75% ┆ 2020-07-18 00:02:56 ┆ 1.5950e9 │
│ max ┆ 2020-07-20 00:03:37 ┆ 1.5952e9 │
└────────────┴────────────────────────────┴───────────────┘Find the average humidity level across all sensors.
df_original.select(pl.col("humidity").mean())
shape: (1, 1)
| humidity |
|---|
| f64 |
| 60.511694 |
Remove unwanted columns
We are only interested in certain columns, the rest of the columns can be removed.
irrelevant_columns = ["co", "lpg", "motion", "smoke", "ts", "light"]
for columnname in irrelevant_columns:
pass
#df = df_original.drop(columnname)
#df_original.head(1)Reorganize the columns
df = df_original.select(['timestamp', 'device', 'temp', 'humidity'])
df = df.sort(by="timestamp")
df.head(1)
shape: (1, 4)
| timestamp | device | temp | humidity |
|---|---|---|---|
| datetime[μs] | str | f64 | f64 |
| 2020-07-12 00:01:34 | "b8:27:eb:bf:9d:51" | 22.7 | 51.0 |
import seaborn as sn
sn.lineplot(df.head(100), x="timestamp", y="temp")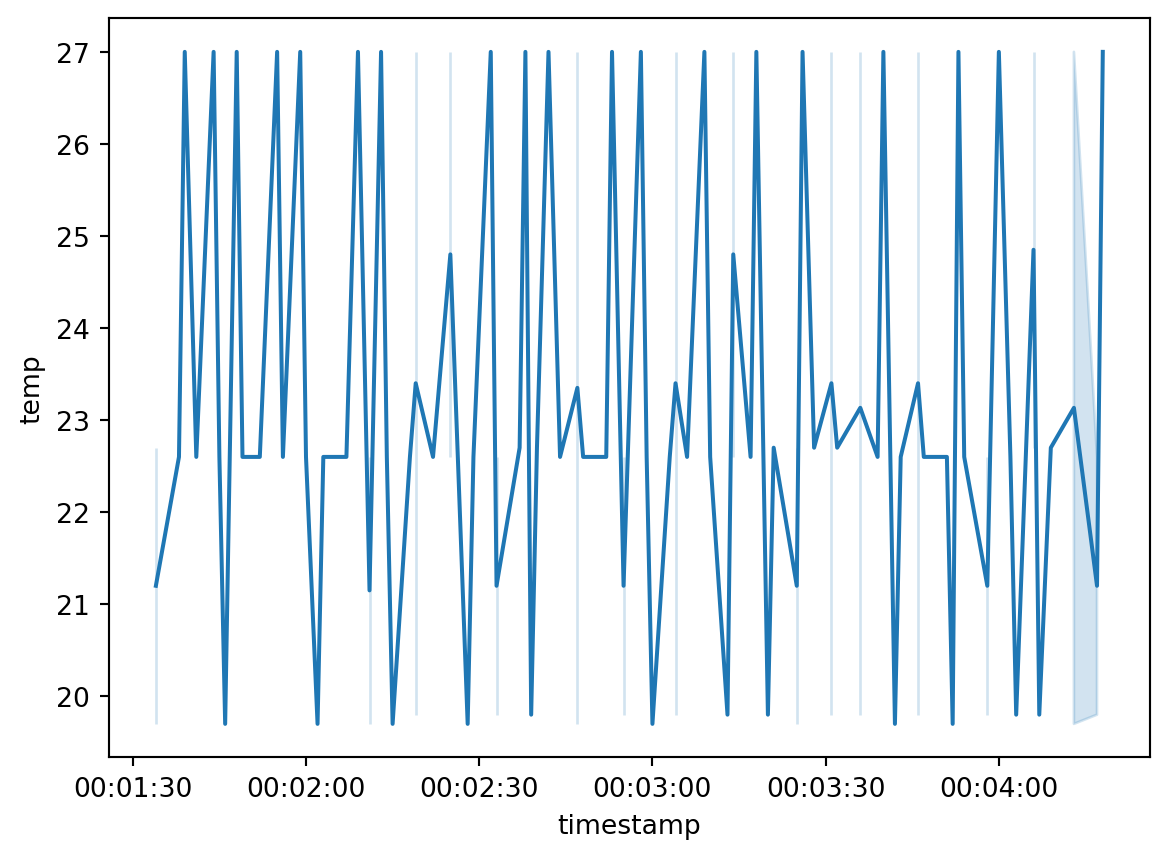
from prophet import Prophet
import pandas as pd
import dyplot
df_device_1 = df.filter(pl.col("device")=="b8:27:eb:bf:9d:51")
df_device_2 = df.filter(pl.col("device")=="00:0f:00:70:91:0a")
df_device_3 = df.filter(pl.col("device")=="1c:bf:ce:15:ec:4d")
def get_forecast_data(df):
device_df = df.select(pl.col(["timestamp", "temp"])).rename({"timestamp":"ds", "temp":"y"})
device_df = device_df.to_pandas()
m = Prophet()
m.fit(device_df)
future = m.make_future_dataframe(periods=4)
forecast = m.predict(future)
forecast[['ds', 'yhat', 'yhat_lower', 'yhat_upper']].tail()
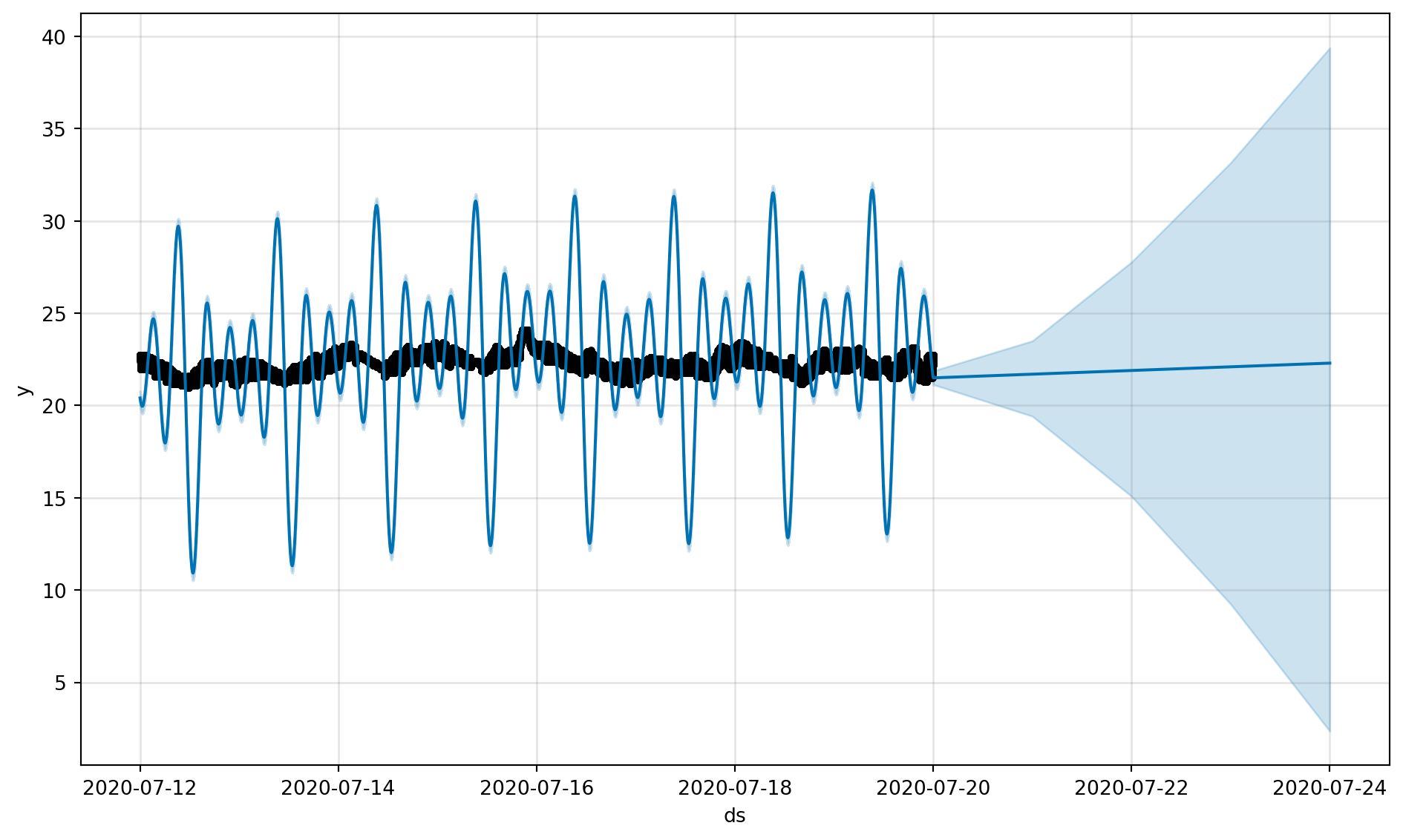
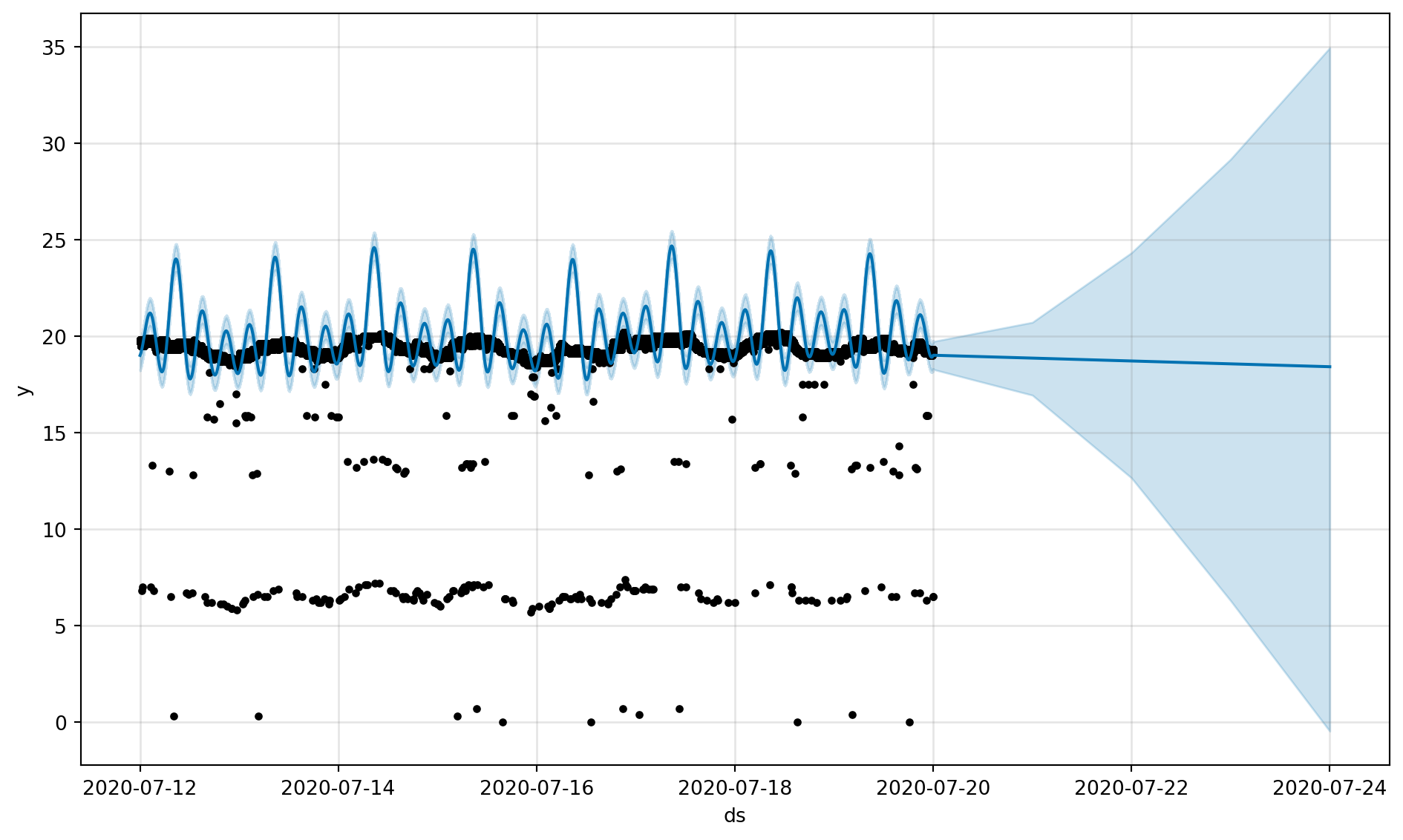
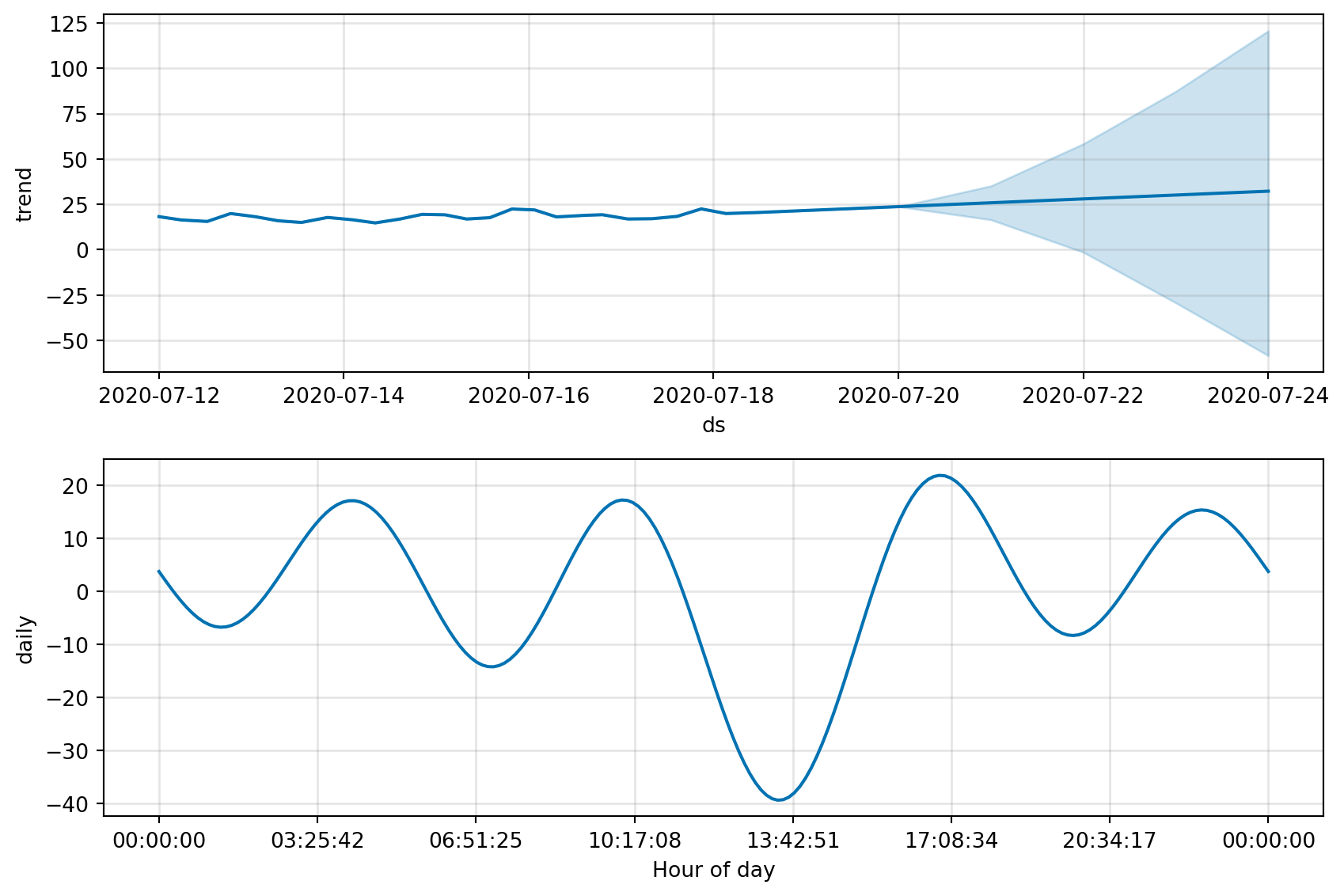
fig = m.plot(forecast)
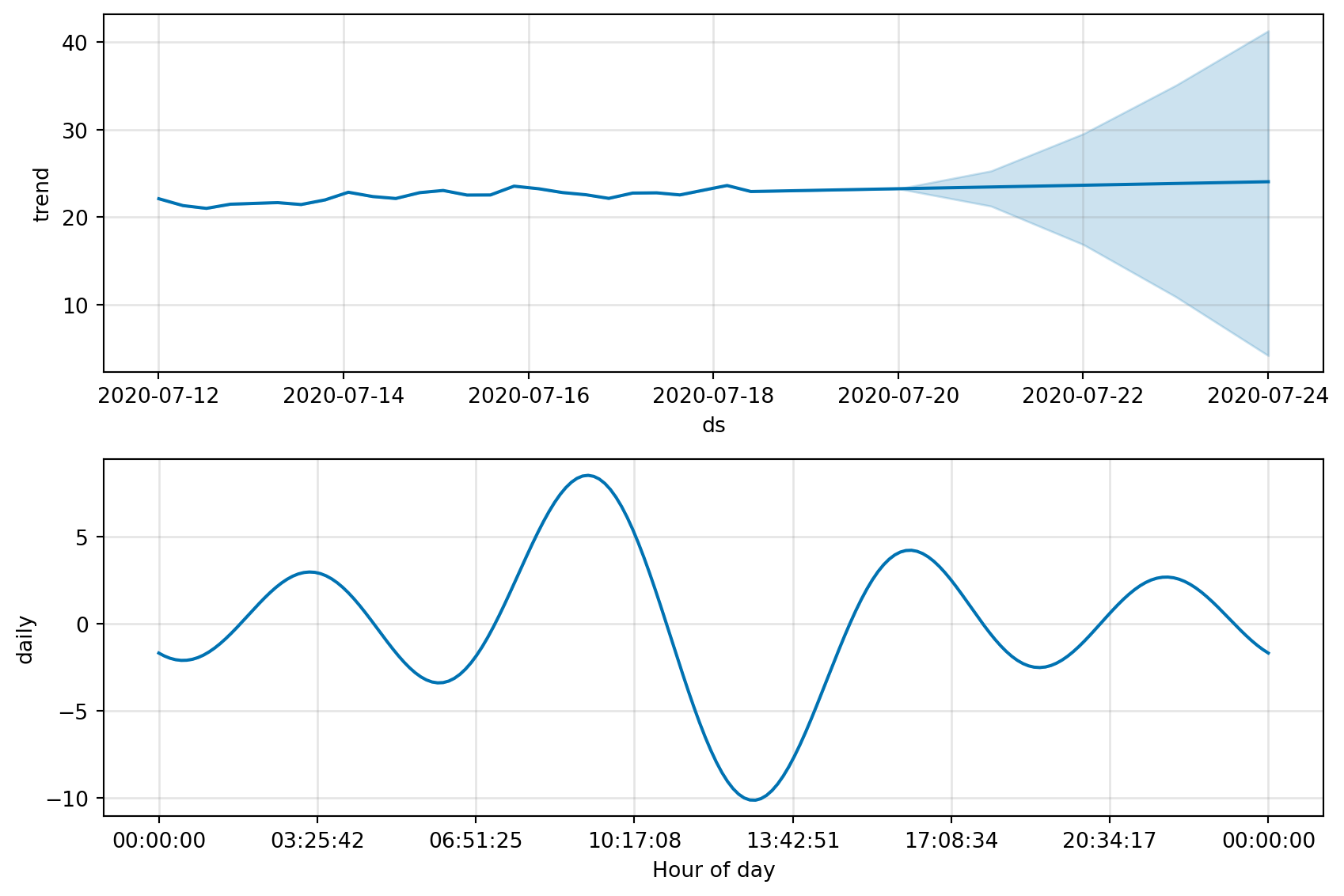
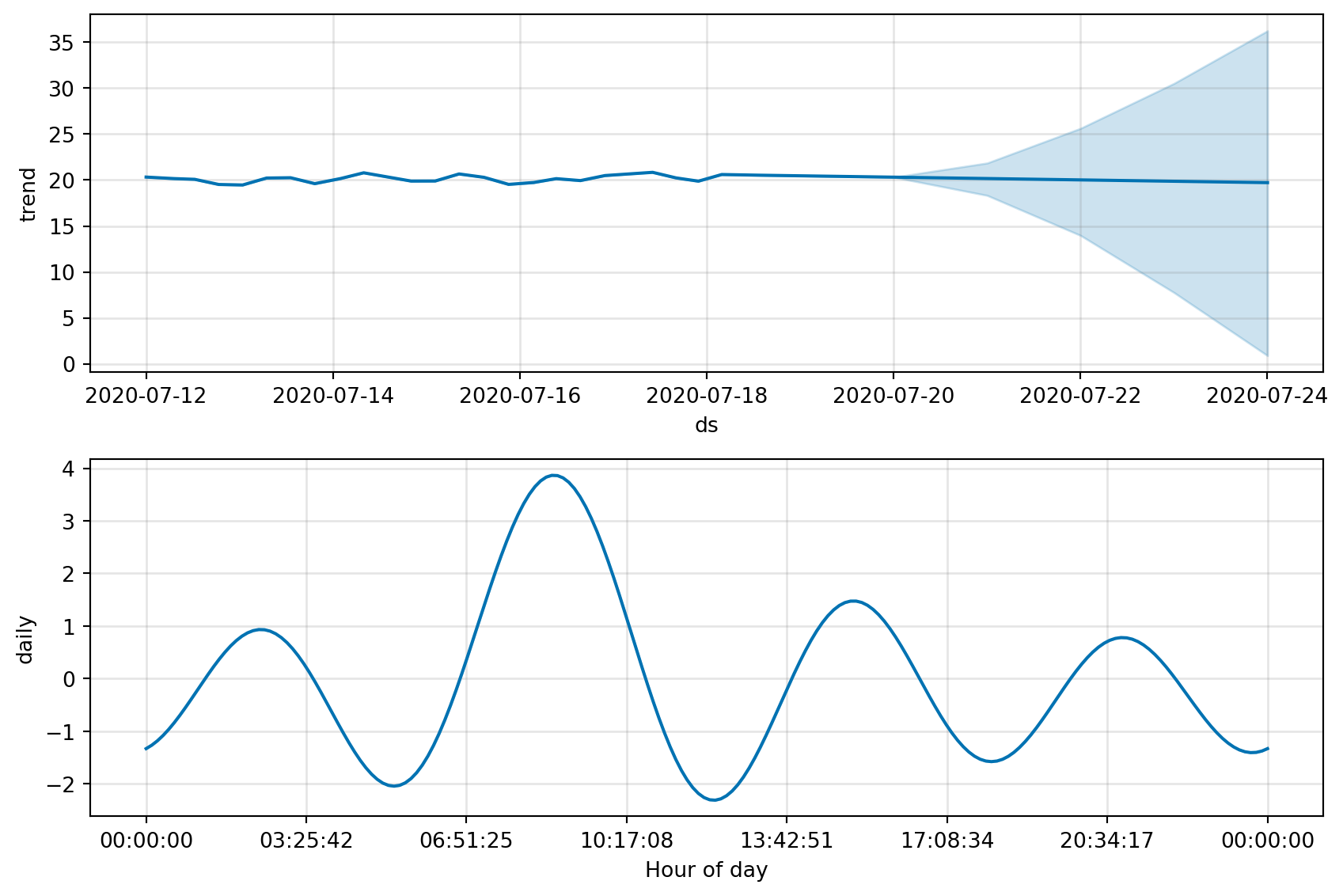
fig_comp = m.plot_components(forecast)
#fig_dyplot = dyplot.prophet(m, forecast)
#fig_dyplot.show()
return fig, fig_comp
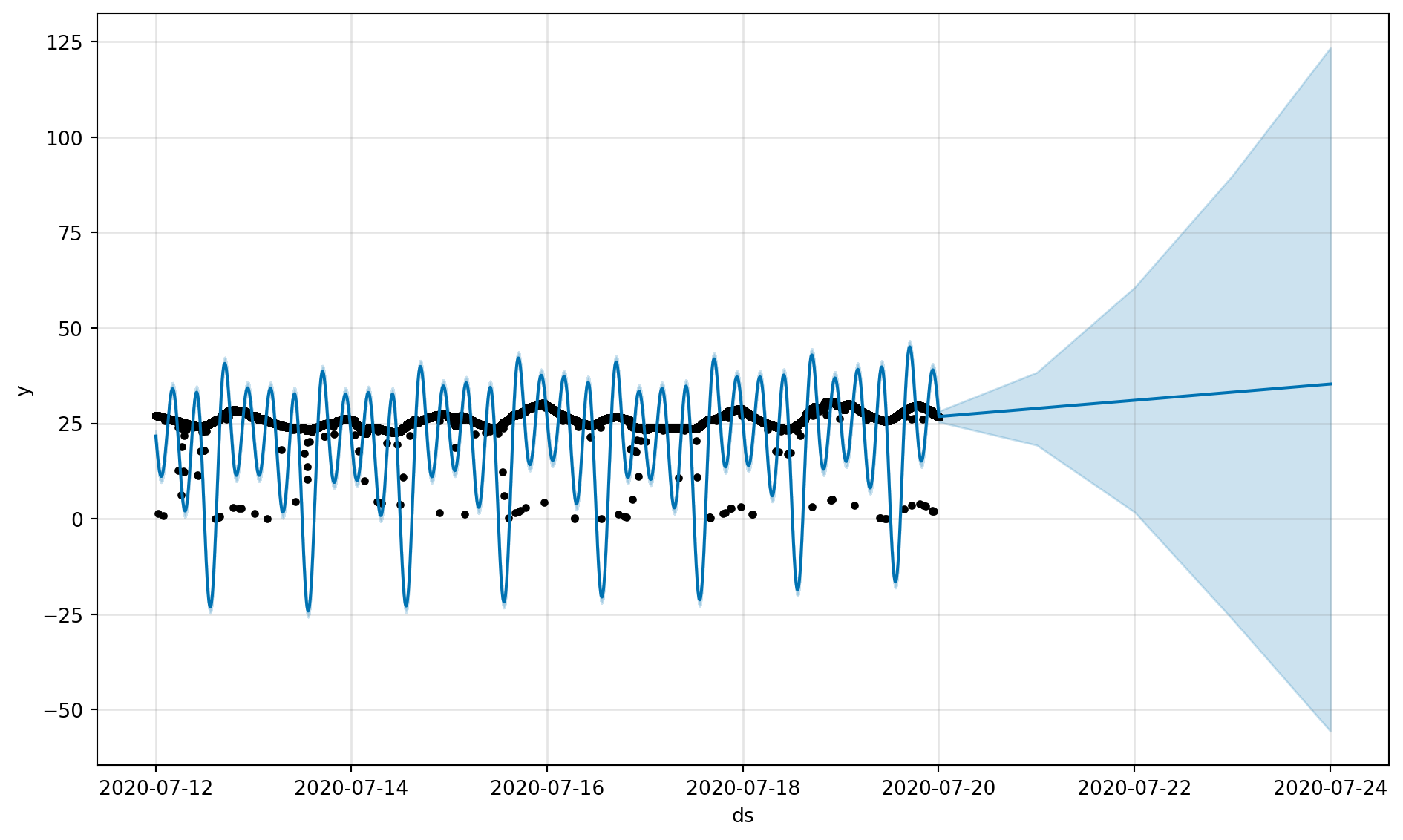
fig, fig_comp = get_forecast_data(df_device_1)
fig, fig_comp = get_forecast_data(df_device_2)
fig, fig_comp = get_forecast_data(df_device_3)21:45:04 - cmdstanpy - INFO - Chain [1] start processing
21:46:24 - cmdstanpy - INFO - Chain [1] done processing
21:46:45 - cmdstanpy - INFO - Chain [1] start processing
21:47:28 - cmdstanpy - INFO - Chain [1] done processing
21:47:41 - cmdstanpy - INFO - Chain [1] start processing
21:48:40 - cmdstanpy - INFO - Chain [1] done processing